Metastasis Research
Our vision is to protect patients from the development of systemic metastases and thus from a lethal course of the disease. For this, we are investigating the mechanisms of systemic spread of cancer in four multidisciplinary research groups. We are developing technologies and models that allow us to study the disease in patients from its earliest beginnings and to understand the evolution of disseminated cancer cells over disease courses and in various metastatic target organs.
AG Klein

Prof. Dr. Christoph Klein
Chair holder
Tel: +49-(0)941-944-6720 or +49-(0)941-944-31220
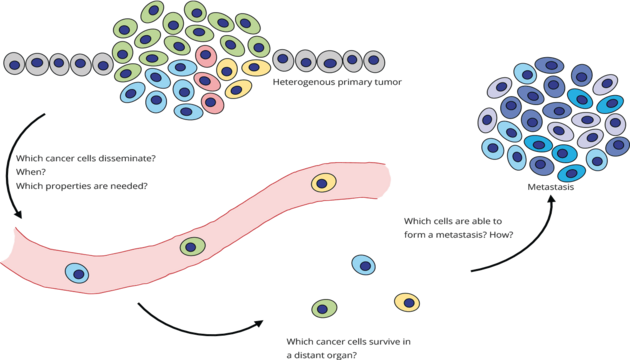
Email: Christoph.Klein@klinik.uni-regensburg.deOur work has shown that dissemination of tumor cells occurs early in the development of a malignant disease and that disseminated cancer cells (DCCs) often have not yet acquired important changes (e.g. mutations) when leaving the primary tumor. Our key research questions are therefore: How do these tumor cells survive at the site of metastasis? How do they acquire the missing and often critical changes? Which role does the microenvironment play in this process? Which cells are the founder cells of metastases?
We investigate these questions by
- generating family trees of individual cancer diseases (cell lineage trees);
- generating gene expression profiles of individual disseminated tumor cells and identifying activated signaling pathways;
- cataloging mutations of DCCs characteristic for the progression stages of metastases;
- generating DCCs from normal cells by genetic modification in order to dissect progression steps;
- performing drug screens with the DCC models.
Our goal is to identify key regulatory mechanisms and rules governing early metastasis formation that can be therapeutically exploited to prevent the onset of lethal metastasis.
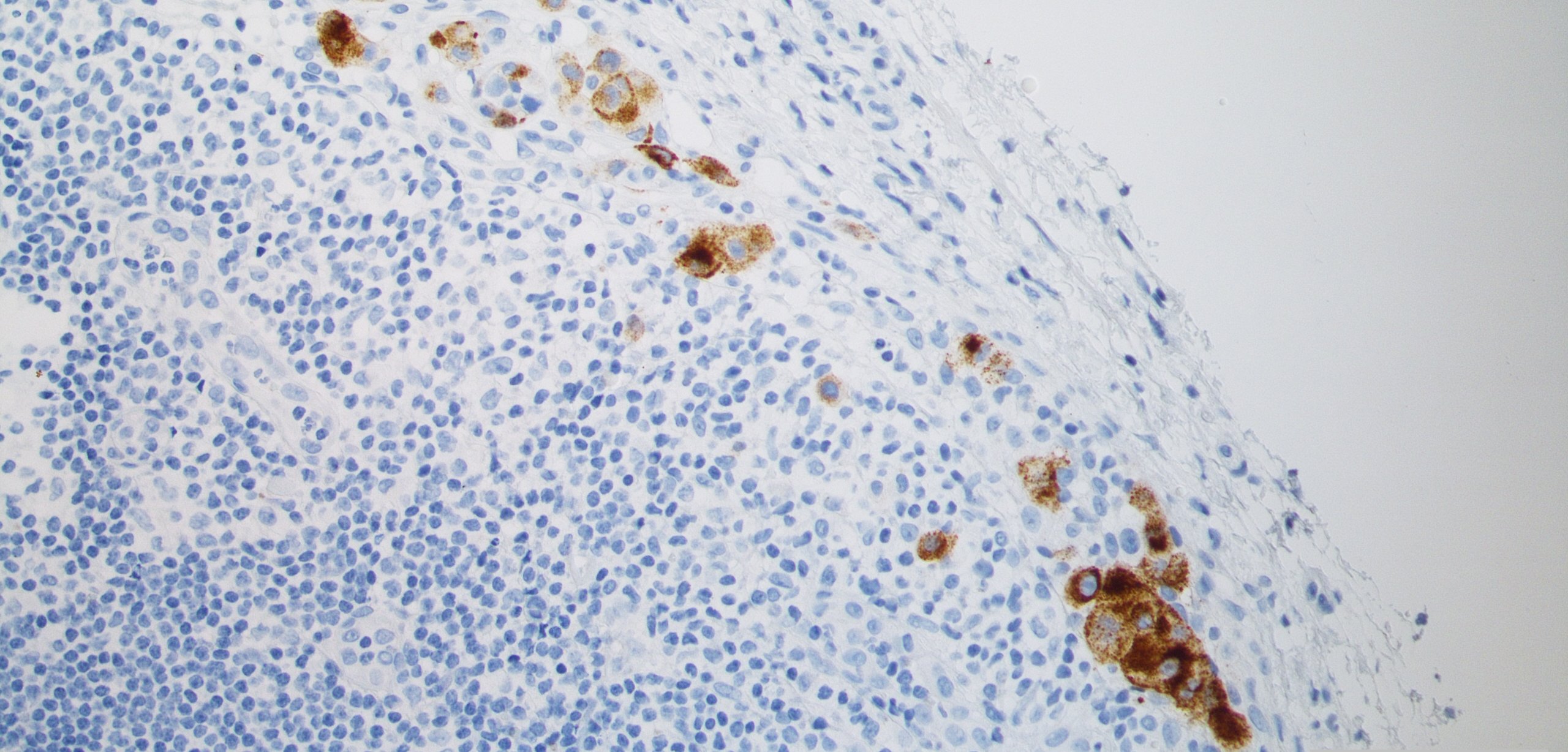
Bioinformatics analysis of DCC (Dr. Huiqin Körkel-Qu)
A special focus of our work is on the development and use of bioinformatics approaches for single cell analysis. DCCs are very rare and their phenotype after dissemination and establishment in the target organ is largely unknown. Since it usually takes months or even decades from DCC detachment to fatal metastasis formation, this time window offers therapeutic options to prevent metastasis by eliminating DCCs early on. To distinguish DCCs from other cells and to understand their molecular mechanisms is a critical step to better conceptualize and prevent metastasis. Our goal is to set up bioinformatics tools that enable reliable DCC identification in a variety of different distant organs including determination of copy number alterations and single nucleotide variants in DCCs.New cell models for the analysis of early systemic cancer (cooperation with Dr. Miodrag Guzvic, Experimental Urology, Chair of Urology, University of Regensburg
We know very little about the phenotype of DCCs upon arrival in the target organ and their response to the new microenvironment. Therefore, we try to understand the biology of DCCs in the context of their microenvironment. We model the metastatic niche in the bone marrow and in lymph nodes in vitro to develop new approaches to study the response of DCCs to novel treatments.
AG Werner-Klein

PD Dr. rer. nat. Melanie Werner-Klein
Group Leader/QM-Specialist
Tel: +49-(0)941-944-11223
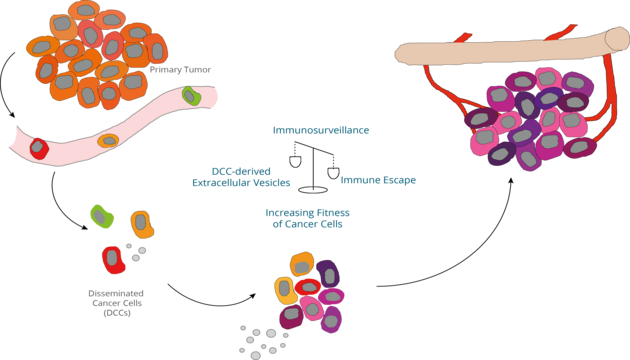
Email: melanie.werner-klein@ukr.deMetastases develop from a handful of systemically spread cancer cells. Disseminated cancer cells (DCCs) originating from epithelial cancers or melanoma can be best identified in hematopoietic organs or lymphatic tissues. Lymph nodes and bone marrow are sites where primary immune response are initiated and immunological memory is kept. Therefore, invading cancer cells need to escape immune defense early on by either suppressing anti-tumoral immune responses or converting the immune-microenvironment into a growth supporting niche. Moreover, DCCs need to acquire important fitness-conferring genetic changes to adapt to the new, local microenvironment and establish a metastatic colony.
To identify cancer cell intrinsic and (immune-) microenvironmental factors operating during the early phase of metastasis formation, we employ latest single cell sequencing technologies, cellular assays and multiparameter flowcytometry using diagnostic patient samples of bone marrow or lymph nodes. To functionally investigate candidate mechanisms, we use patient-derived xenografts of DCCs and immune/tumor humanized mouse models.
We are specifically interested in the DCC-immune cell interactions for the metastasis success of DCCs. This includes inter-cellular communication via extracellular vesicles, evolution of neo-antigens and the role of inflammatory cytokine signals.
AG Czyz

Dr. Czyz Zbigniew
Group LeaderTelefon:+49 941 944-11224
E-Mail: zbigniew.czyz@ukr.deSingle-cell sequencing technologies enable to deconvolve the complexity of cell populations with an unprecedented level of resolution. In cancer research single-cell sequencing approaches are invaluable to assess both inter- and intratumor heterogeneity, but also allow to uncover and track evolutionary processes.
Cancer cells during minimal residual disease after therapy or during the early phase of the disease are characterized by an extremely low abundance and high level of genetic heterogeneity. This biological framework renders their analysis technically challenging and requires highly accurate, sensitive and robust analytical procedures.
Single-cell molecular profiling assays initially focused on the analysis of a single layer of gene regulatory network, that is genome, epigenome, transcriptome and proteome. But sampling of just one molecular type from individual cells provides only an incomplete picture of gene expression dynamics and cellular states determined by a complex interplay of different molecules. Therefore, the goal of this team is to develop and implement novel single-cell multi-omics technologies enabling a highly accurate and parallel analysis of several molecular levels of disseminated or circulating cancer cells. This innovative approach will pave the way for better understanding of processes operating during early systemic cancer or minimal residual disease and elucidate new therapeutic options.




